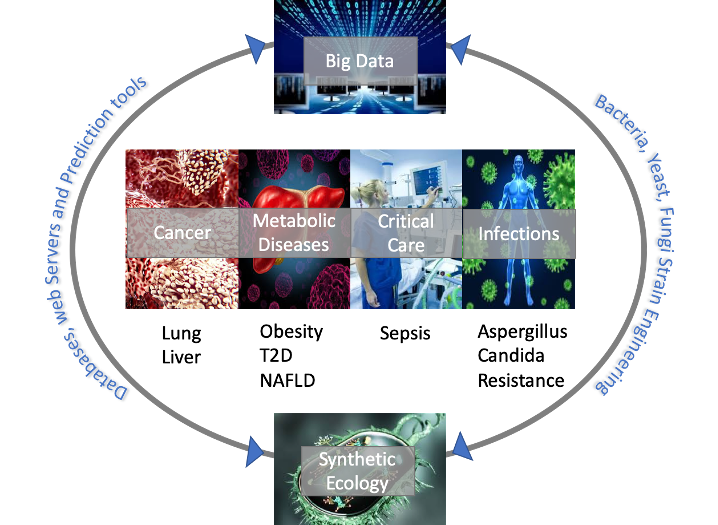
Our research is highly collaborative and explores the role of the microbiome in globally significant diseases for delivering novel patient-centric therapies. We are studying several diseases that fall under the following broad categories: metabolic diseases, infection, cancer and critical care. In order to move from correlative to causative evidence between shifts in the microbiome and pathophysiology of diseases my group has secured access to cross-sectional studies, prospective studies and randomized controlled intervention studies in disease subjects and a combination of emerging new technologies, including transplantation of human fecal microbiota to germ free and evaluation of single species and microbial consortia in stem cell lines and artificial gut systems. We are also particularly interested on the dynamics between host and the associated microbes therefore we are working on algorithm development for generating probabilistic frameworks for learning persistence and colonization probabilities of microbiome therapeutics and microbial engineering. We also combine clinical data, host mutation patterns, microbiome, mycobiome and phageome data to create a comprehensive view of different diseases, identify subtypes and predict response treatment.